Editorauthors are masked to the peer review process and editorial decision-making of their own work and are not able to access this work in the online manuscript submission system. Each pure protein has a unique extinction coefficient.

Measuring Protein Concentration With Uv Absorbance And Bradford Assay The Bumbling Biochemist

Protein Concentration Using Excel 2016 Youtube
1
Salt concentration 100200 mM.
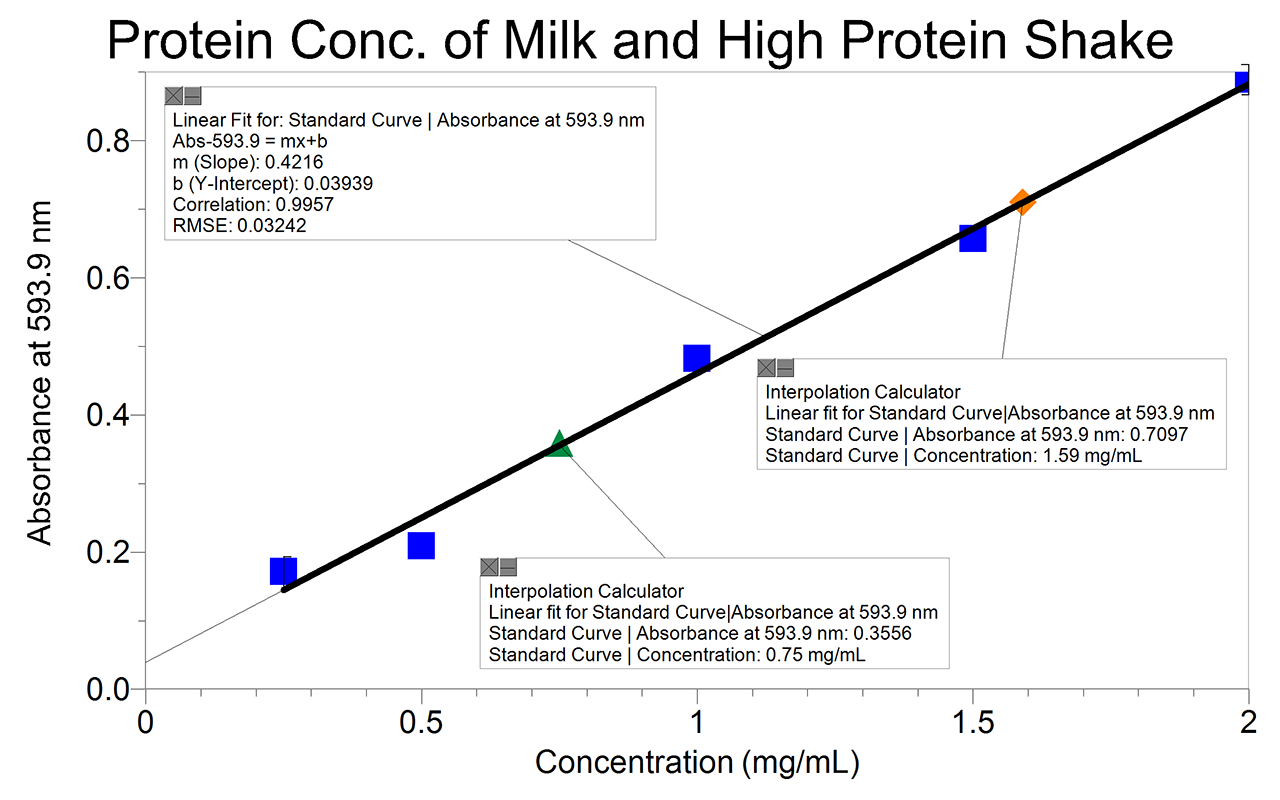
Protein concentration absorbance calculator. This translates into 1125 mgml of BSA protein or 37500 nguL of dsDNA. Molar concentration also called molarity amount concentration or substance concentration is a measure of the concentration of a chemical species in particular of a solute in a solution in terms of amount of substance per unit volume of solution. In chemistry the most commonly used unit for molarity is the number of moles per liter having the unit symbol molL or moldm 3 in SI unit.
The hemagglutination assay HA is a common non-fluorescence protein quantification assay specific for influenzaIt relies on the fact that hemagglutinin a surface protein of influenza viruses agglutinates red blood cells ie. Recombinant protein is a manipulated form of protein which is generated in various ways to produce large quantities of proteins modify gene sequences and manufacture useful commercial productsThe formation of recombinant protein is carried out in specialized vehicles known as vectors. Calculate nucleic acid concentration from absorbance and vice versa.
If the mean concentration of Cryptosporidium exceeds. Steady State and Lifetime Benchtop Spectrofluorometer. The purified protein can be dialyzed and concentrated to obtain the target band with darker color indicating that the concentration is more effective and the protein concentration is.
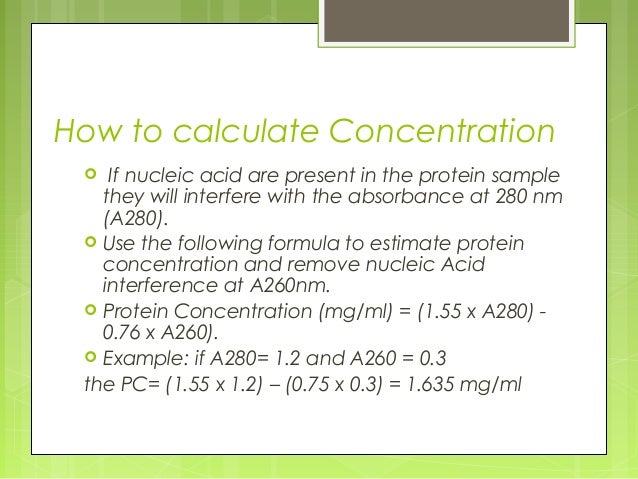
022 x 10 23 units called. The FluoroMax family with its unique all reflective optics and photon counting was the first to bring the sensitivity of a modular fluorometer to a tabletop fluorescence instrument. Using the absorbance at 280nm A280 protein concentration c is calculated using the Beer-Lambert equation A 280 c ε b ε is the wavelength-dependent protein extinction coefficient b is the pathlength.
Using the medium the cell suspension is diluted to a concentration gradient usually requiring 5-7 concentration gradients several replicate wells per. Let me get the calculator back. 1 iv Use Equation 2 to calculate the number of moles of iodine I 2 that reacted with The volume of gas collected and the gas laws can be used to calculate the number of moles of gas collected.
This ultra high absorbance capability is achieved by using a remarkably small pathlength enabled by our SmartPath Technology. The FluoroMax series represents HORIBAs industry-leading fluorometer performance in a convenient affordable easy-to-use benchtop model. O-l Appendix P Validation Protocol Calculator Tool P-l UV.
Protein concentration 110 mgml. Storage at 4C with 10 mM sodium azide or 0005 merthiolate is usually safe over long periods but occasionally proteolytic breakdown occurs. The gene coding for this enzyme was.
Bioinformatics analysis of the complete transcriptome of Fasciola hepatica identified a total of ten putative carboxylesterase transcripts including a 3146 bp mRNA transcript coding a 2205 bp open reading frame that translates into a protein of 735 amino acids resulting in a predicted protein mass of 835 kDa and a putative carboxylesterase B enzyme. The cell counting plate counts the number of cells in the cell suspension. Conversely you can calculate the molarity of a nucleic acid solution prepared by dissolving a certain amount of it in a specified volume of a solvent.
Causes red blood cells to clump togetherIn this assay dilutions of an influenza sample are incubated with a 1 erythrocyte solution for one hour and the virus. However the protein concentration when diluted by assay reagent is almost certainly not the value of interest. The DS-11 can measure samples with absorbance values as high as 750 absorbance units at a 1 cm equivalent path length.
24 Scattering of Light 2-5 Figure 25 Structure of DNA and Nucleotide Sequences Within DNA 2-6 Figure 26 UV Absorbance of Nucleotides and Nucleic Acid at pH 7 2-7 Figure 27. 260230 The 260230 ratio is a value that reflects how pure the sample is from salts and. Because the original standard was 1000 µgmL the test sample that produced the.
Calculate the mass or volume required to prepare a nucleic acid solution of specified molar concentration. B Calculate the number of moles and the mass of MgCO3 in the sample and hence deduce the percentage of MgCO3 in the sample. Cells tissue and re-extract the whole sample.
Storage at 20C in PBS in 50 glycerol is usually safe over long periods. 50 glycerol does not freeze at 20C. AJOGs Editors have active research programs and on occasion publish work in the Journal.
Ac control well absorbance absorbance of wells containing cells medium and CCK8. Instead one wants to know the protein concentration of the original test sample. Alternatively if the sample has a low concentration of nucleic acids to start with it may be best to go back to the source eg.
Making a standard curve. After the protein was purified by the nickel affinity column the target protein with the expected size was obtained Fig.
Protein Det

Use The Spectrovis Plus To Study Proteins

Absorbance Based Methods For Protein Quantification Bmg Labtech

Photometric Quantification Of Proteins In Aqueous Solutions Via Uv Vis Spectroscopy Eppendorf Handling Solutions
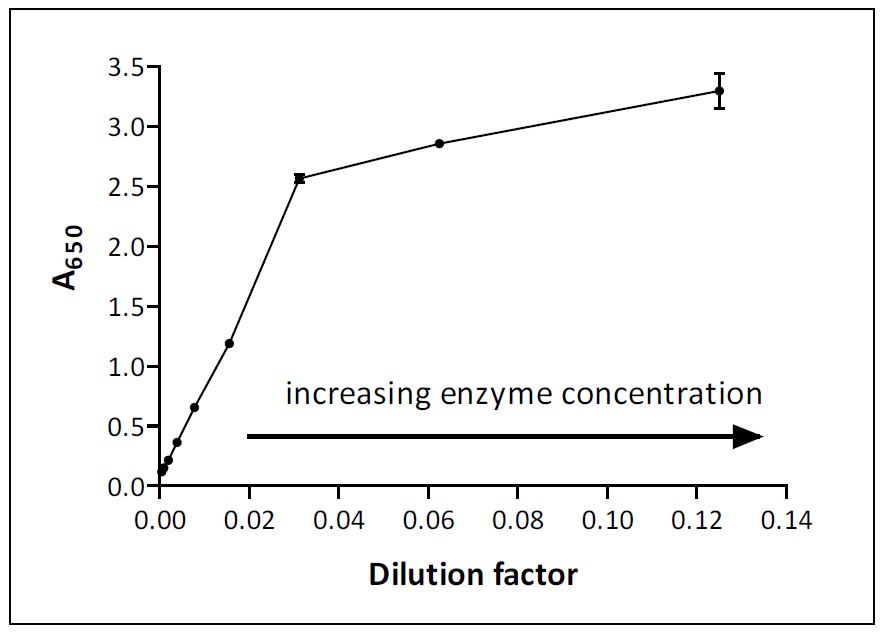
Guide To Enzyme Unit Definitions And Assay Design Biomol Blog Resources Biomol Gmbh Life Science Shop
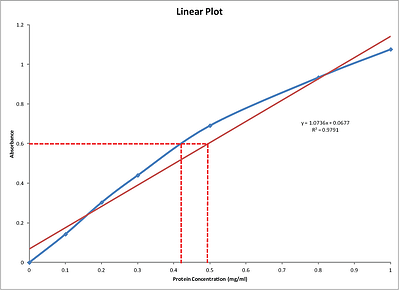
Bradford Protein Assay Calculation Of An Unknown Standard
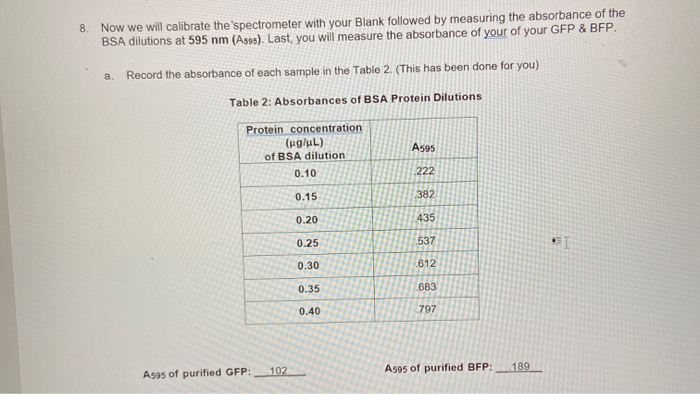
Solved 6 Calculate The Protein Concentrations Of The Chegg Com
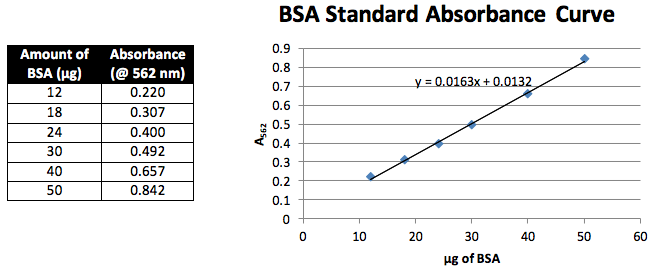
Bca Protein Concentration Assay Odinity
